ddPCR: Exploring the Progress of PCR Technology and Novel Applications

Polymerase Chain Reaction (PCR) is an in vitro technique that amplifies DNA, generating several millions of copies of a specific segment of DNA from a minute amount of starting material. From humble beginnings, there have been several evolutions of this technique. Today, ddpCR is being embraced by both academics and pharmaceutical companies. Additionally, it is making headlines for innovative new use cases, including calculating the severity of SarsCov2.
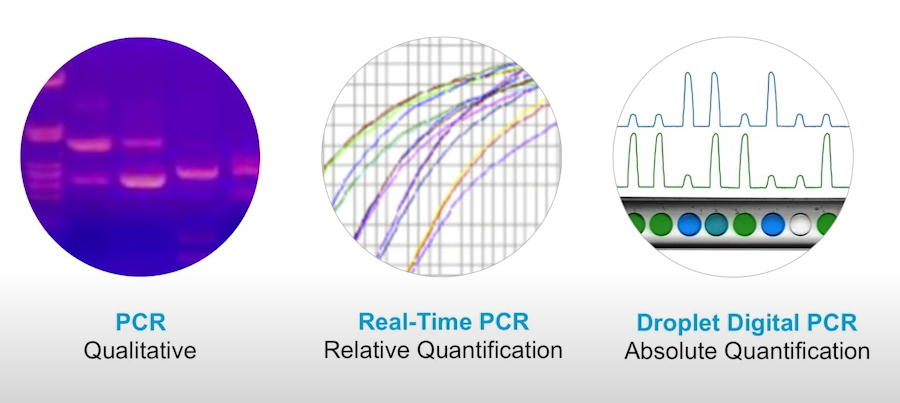
PCR: Progress and Rationale for Use
The story of modern PCR begins in 1976 with the isolation of Taq DNA polymerase from the thermophilic bacterium Thermus aquaticus. Its isolation meant that molecular biologists now had a thermostable enzyme that was capable of repeat PCR cycling without the need to add fresh DNA polymerase after each cycle.
In 1989 Kary Mullis and the Cetus Corporation commercialized PCR for the first time. Taq DNA polymerase was an instant success, even winning Science magazines 'molecule of the year.”
Although these developments represented significant progress, Taq DNA polymerase wasn't perfect. It was unstable at high temperatures and error prone and had difficulty amplifying DNA rich in GC content or with strong secondary structure. These factors played a role in the stunted development of PCR early on, particularly in applications that required high specificity and reliability.
Real time PCR
In the early 1990’s Higuchi et al recognized the process of PCR could be monitored by addition of a fluorescent label that binds to the accumulating PCR product. As the concentration of PCR product increases, the intensity of the fluorescence signal also increases. This discovery paved the way for modern quantitative real-time PCR (qPCR). In current qPCR technology, these fluorescence signals are generated by inclusion of either fluorescent DNA-binding dyes or oligonucleotide probes.
Digital PCR
The readout of digital PCR differs to real time PCR. In real time qPCR you measure the threshold cycle, while a ddpcr readout is either positive or negative. By counting the number of droplets that include the target (positive) and the number without the target (negative) and applying Poisson statistics you can calculate the copies per microliter: the concentration. To make an estimation of concentration in digital PCR, you need to have at least some number of negative droplets and some number of positive droplets
Digital polymerase chain reaction technologies (dPCR) have a range of benefits compared to conventional PCR methods. Nevertheless, they have a few disadvantages as well. Namely, standard dPCR techniques lack practicality and aren’t scalable. Samples are diluted serially and manually, which can lead to pipetting errors. If partitioning is performed via a chip-based system, it is time-consuming and complex to manage the fluidics schemes required for it.
Droplet Digital PCR
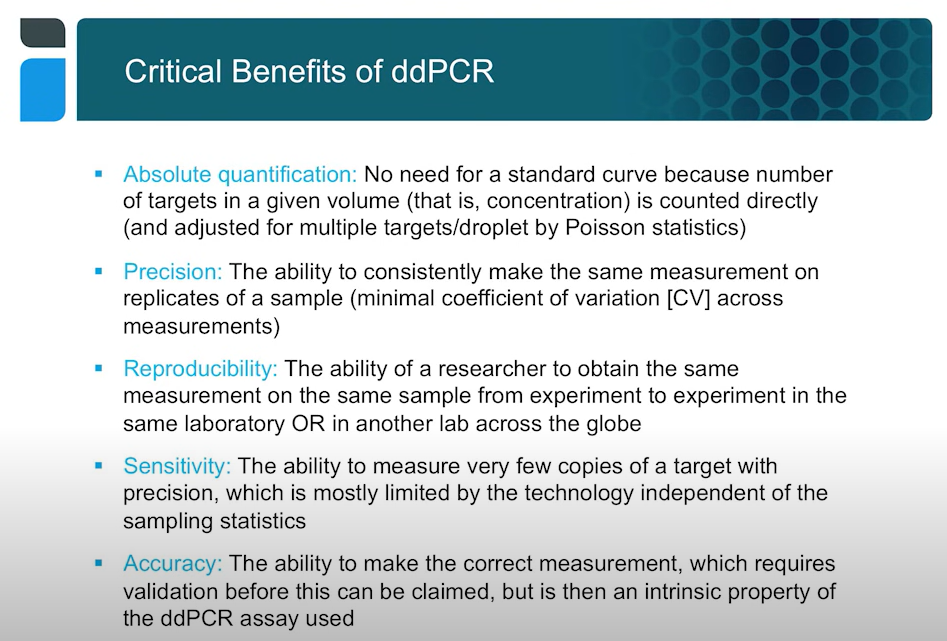
ddPCR does not come with the same issues. The partitioning process is done via a droplet generator that makes it incredibly easy to separate each sample well into twenty thousand droplets. If a good system is used, all the droplets are uniform in size and shape, meaning the results are robust and reproducible.
Unlike real time PCR there is no need for a standard curve, which eliminates some of the difficulties that creates. A sample may have matrix components can alter the rate at which your fluorescence rises in a real time reaction and lead to uncertainty. Having reliable standards that behave the same way as your experimental sample is important.
Another important feature is reproducibility, both for experiments in the same lab and across the world. Reproducibly sensitivity is also a key measure. Greater precision leads to greater sensitivity. Where a measurement has X amount of target or half that amount, that you can discriminate those from each other more readily when you have very little uncertainty about each of those two measurements.
Case Study: Calculating Sars-Cov2 Severity
Researchers at the University of Paris have been using ddPCR to understand Sars-Cov2 severity. They found that the positive detection of SARSCoV-2 RNAaemia by ultrasensitive, droplet digital PCR was correlated with future clinical deterioration, intubation, and death.
SARSCoV-2 RNAaemia detection has proved invaluable for predicting patient outcomes. Clinical recovery was correlated with the negative action of the SARSCoV-2 RNAaemia, detected and quantified by droplet based digital PCR. They confirmed in this study on almost 130 patients suffering from COVID-19, finding that SARSCoV-2 RNAaemia was closely correlated with clinical severity and ventilation needs. The plasmatic RNA concentrations used significantly correlated with the clinical severity, more than the viral load. Research is now underway to examine if this could also be the case with other infectious diseases.
Final Thoughts & Conclusion
Droplet digital PCR (ddPCR) is a recently introduced technology that has proved to be more sensitive, accurate, and reproducible than previous methods. While it is still in its infancy, ddPCR has already proven its worth during the COVID-19 pandemic and work is underway for a wide array of use cases. For the latest information on ddPCR consider joining us for our upcoming NextGen Omics: UK event, which will feature a dedicated PCR track and multiple presentations from key opinion leaders.
Related Resources