Single-Cell Transcriptomics and Implications for COVID-19 Severity
Presented by: Zhongming Zhao, Chair Professor for Precision Health at the University of Texas Health Science Centre, Houston
Edited by: Ben Norris
The Coronavirus disease 2019 (COVID-19) pandemic has already infected hundreds of millions of people, causing numerous health problems and many millions of deaths worldwide. This disease is caused by SARS-CoV-2 which is a single-strand RNA virus with 30 kilobases and moderate mutation rate among all other viruses. Zhongming Zhao, Chair Professor for Precision Health at the University of Texas Health Science Centre, explained that some of the genomic data from COVID-19 cases with different severity presented a unique opportunity when analysed through single-cell transcriptomics. His laboratory was able to assess the genomic and genetic changes of genes involved in virus infection across the patients subject to the severity of their response to SARS-CoV-2 virus.
Coronavirus Architecture and Lung Biopsies
Zhao opened his presentation at Oxford Global’s NextGen Omics US event in March 2022 by highlighting the architecture of the SARS-CoV-2 transcriptome – better known to most of the world as the virus that causes COVID-19. “The virus is everywhere, basically,” Zhao said. “It’s a positive sense, single stranded RNA virus, which is very important because we deal with many kinds of viral research. So far we don’t have much information on how this virus can be prevented, and no evidence in its ability to integrate into the host genome.”
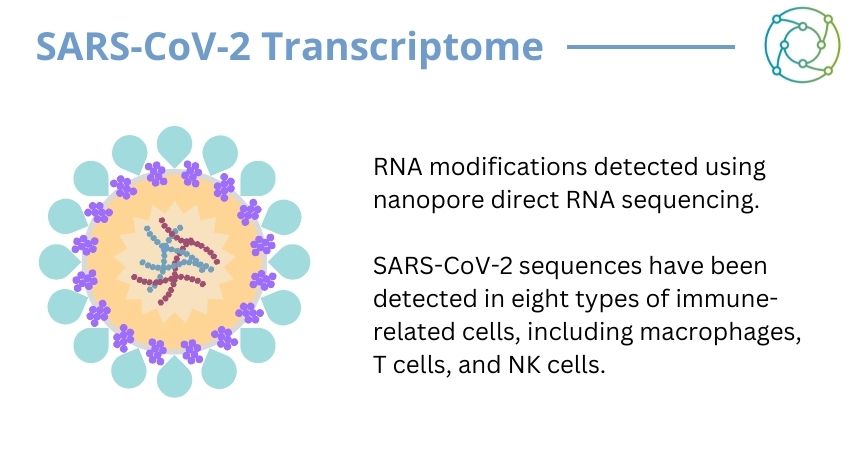
Zhao explained that his laboratory has run analysis on the SARS-CoV-2 transcriptome, talking the audience through his familiarity with some of the spike membranes and coordinates along its 30 kilobase length. In addition, Zhao also investigated 65 COVID-19 scRNA-sequence datasets through single-cell RNA sequencing. “They are a set of very precious datasets for the real transcriptome and genome analysis,” Zhao explained.
One of the datasets used to explore this aspect of the virus's functionality involved bronchoalveolar immune cells obtained from patient lung biopsies. “This is very unique because such tissue is usually very difficult to obtain,” said Zhao. His belief was that a certain gene mutation had a higher likelihood to occur more frequently in samples from severe cases of Coronavirus versus more moderate cases. “Very quickly I formed a team,” Zhao continued. Here he mentioned a former Postdoc, Teng Liu, PhD, and Bingliang Fang, in Immunology — “we worked together to reanalyse the single-cell transcriptomic (scRNA-seq) data from patients.”
Single-Cell Transcriptomics and Major RNA Functions
The investigation led by Zhao analysed the cell distribution for all SARS-CoV-2 infected bronchoalveolar lavage fluid (BALF) samples, with detection of virus sequences in different cell types including macrophages. From this research, the virus was shown to be detected in eight cell types: epithelial cells and macrophages exhibited infected cells in both moderate and severe cases. “We examined the gene expression of the Coronavirus in these genes, and found primary high expression in the severe group,” said Zhao. “It was important to further validate them as the sample size is still relatively small.”
- Novel Gene Editing Approaches and Techniques Alongside CRISPR-Cas9
- Tomorrow's Healthcare: Pioneering Approaches to Gene Therapy
- Current Bioinformatics Trends in Single-Cell Data Analysis: Good Practice Makes Perfect
Following the publication of this paper, Zhao and his team looked specifically at the genes in different groups. They observed that major viral RNA fusions were present in infected cells, and that the incidence and behaviour of fusion events could predict patient symptoms in moderate or severe cases of COVID-19. With analysis of the transcription regulatory sequence of the viral leader sequence, Zhao found that gene incidence was dependent on RNA fusions.
Zhao first replicated these features with his team and then observed a new fusion event linked to severity. “Interestingly, we find another starter location at the 1073 position where we’ve observed many more gene fusion events,” he said. Subsequently, the focus was on finding whether these gene fusion events exhibited a particular severity in gene sequencing. The synopsis of deadly fusion reads has a low survivability in patients, and the clinical outcome is poor.
Gene Expression Adjacent to Severe COVID-19 Cases
Having found that BALF scRNA-seq data was detected in SARS-CoV-2 sequences in eight immune-related cell types, Zhao added that his team first reported that SARS-CoV-2 reads were detected in T cells and NK cells. Transcription regulatory sequences (TRS) of the viral leader sequence-independent fusions with a characteristic 5’ joint point at a specific position along the SARS-CoV-2 genome could be detected exclusively in patients who suffered severe COVID-19 cases. This suggested its potential as an indicator of clinical outcomes for patients infected with the virus.
From this data, Zhao observed that some demographics are at a higher risk of infection from COVID-19. He put together a subsequent study identifying the possible viral mechanisms that may function as a risk to safety. This study identified the genetically regulated gene expression (GREx) for the COVID-19 severity phenotype. The research team observed their features at the single-cell transcriptome level and elucidating the possible mechanisms in the risk to safety posed by COVID-19. The study, carried out through single-cell transcriptomics, monitored the activity and function of CXCR6, a protein coding gene.
Findings and Future Implications in Single-Cell Transcriptomics
“We developed a pipeline with these components for the three genetic markers at the locus,” explained Zhao. “Markers were put in together to study the formation, with DEG analysis for CXCR6 among both moderate and severe patients.” CXCR6 was observed to occur at significantly lower levels of expression in severe COVID-19 patients. In a severe group of COVID-19 patients, the variants in the host genome exhibit a lower expression linked to the residual memory T cells. These are more active and cause additional tissue damage.
One of the most important findings for Zhao was the observation that CXCR6 expression was significantly lower in severe COVID-19 patients than in moderate patients. As the related evidence, a previous mouse study reported the absence of CXCR6 had significantly decreased airway CD8 Trm cells due to their altered trafficking within the lung.
Rounding off, Zhao added that one area his lab had focused on in the last three to four years was single-cell transcriptomics, with a focus on scRNA-seq analysis workflows and computational methods. In terms of CXCR6 expression, the protein was found to have low expression in severe COVID-19 cases, but high expression or over-expression in more moderate cases. For Zhao, the next step is to obtain and analyse further data to build up a more cohesive understanding of the interoperability between SARS-CoV-2 and CXCR6 expression.
Want to read more about the single-cell imaging and treatment of diseases such as COVID-19? Head to our Omics and Biologics portals for new insights from the industry’s best and brightest. If you’d like to register your interest in our upcoming NextGen Omics UK conference, visit our event website.