Are Genomic Technologies the Key to Unlock Precision Immunotherapies?
Since the sequencing of the human genome, scientists have speculated, experimented, and ultimately developed ways of using the growing field of genomics toward therapeutic endpoints. Of course, cancer has always been considered a disease with considerable potential for applying genomic technologies to this goal due to its origin in genomic mutations.
As there is so much variety in the mutations that cause cancer, scientists have endeavoured to look deeper into the nucleus of our cells to produce the best strategies for treating malignancies. Therefore, treating cancer in a more efficacious way will often require a more personalised approach—and that’s where genomic technologies play their role.
“Treating cancer in a more efficacious way will often require a more personalised approach…”
Immunotherapy is one such discipline that is moving in this direction, but there are challenges for this approach. One of those challenges is that inspecting the genome of a tumour cell is often not enough to tell scientists precisely what is going wrong with that cell. Multiple genes may be mutated, all contributing to the overall malignancy of its genome. The tumour cell’s genome needs to be looked at within the context of healthy populations to understand this.
The Advantages and Challenges of Cell Subtyping
Sequencing allows scientists to look at the genetic code in detail and place it within this context. Dr Miao-Ping Chien of Erasmus University Medical Center and UFO Biosciences, who participated in Oxford Global’s Genomic Technologies workshop, is working on the next generation of sequencing techniques. Dr Chien presented her team’s latest research in using cell subtyping to better understand how to treat cancer indications. Cell subtypes are especially helpful, as “well-defined cell subtypes can help robustly identify target genes”.
A problem with traditional sequencing methods such as bulk RNA is that helpful information about specific genes can hide amongst the averages that bulk sequencing provides. Dr Chien asks us to imagine an example of a healthy and diseased population. She explains that if a gene (“gene X”) only causes disease in specific cell types and subtypes, bulk sequencing may draw the misleading conclusion that gene X expression is comparable in the healthy and diseased populations. In actual fact, to see the real difference, you may need to look at the particular gene expression in specific cell types and subtypes.
For example, overall, there may be similar gene X expression from a bulk analysis, but the expression profile for specifically CD8+ T-cells may present a meaningful difference. Without this higher resolution sequencing, missing that meaningful difference would mean missing a target gene that could have significant therapeutic potential.
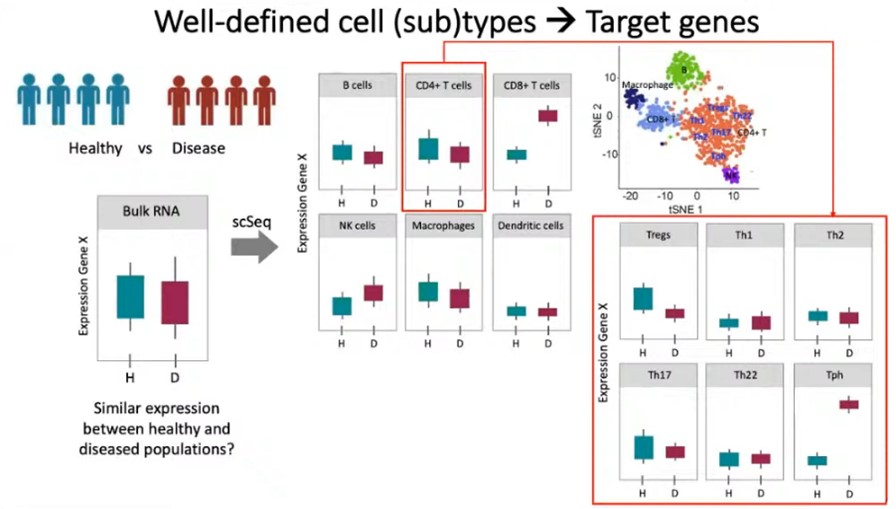
Some methods have been developed to gather this kind of information; Dr Chien uses the example of antibody assisted cell subtyping but explains that it is strongly contingent on having robust and available biomarkers. “There are some immune subtypes where there are no clear or robust biomarkers that we can use to subtype cells…and in cancer cells, this is even more of a problem.” Tumour cells are known to be very heterogeneous, and often that means that you can’t calculate how many subtypes there are in a given sample.
“There are some immune subtypes where there are no clear or robust biomarkers that we can use to subtype cells…and in cancer cells, this is even more of a problem.”
Dr Chien then outlines her approach to this problem, a simple yet ingenious one. She mentioned that although there is so much variation in mutation and heterogeneity in tumour cells, their functional phenotype is often very similar: the phenotypes that are in the interest of their survival. Therefore, Dr Chien and UFO Bioscience suggest subtyping tumour cells based on their functional phenotypes rather than unreliable or unavailable biomarkers. Once these subtypes have been determined, they can work backwards to understand the causative driving mechanism behind their functional phenotype.
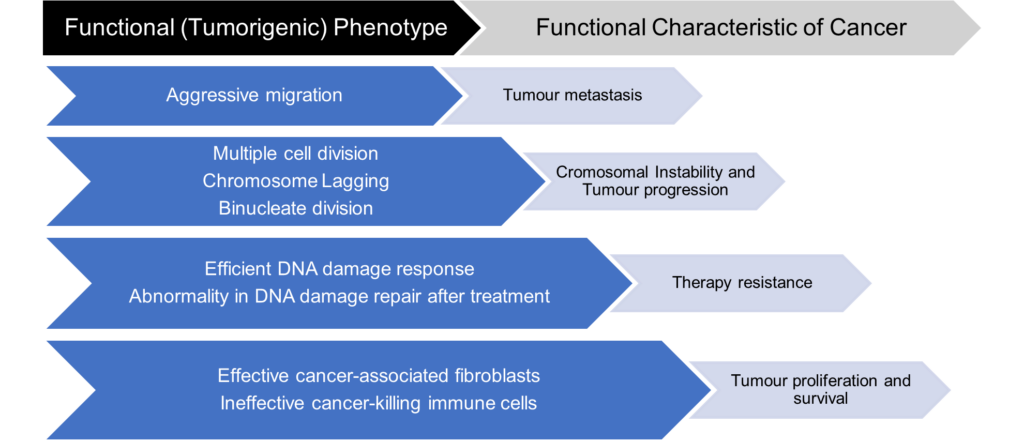
UFO Bioscience developed its enabling technology with this idea in mind. Their custom-built ultrawide field-of-view optical (UFO) microscope can perform high-throughput single-cell screening on up to 100 000 cells with an impressively high spatial and temporal resolution. The vast yielded dataset is then analysed via a real-time algorithm that immediately identifies the phenotypes of interest to be isolated. The isolation process is embedded directly into their technology, isolating as many cells as needed. The last step in this pipeline is to subject them to downstream single-cell sequencing and directly link those genotypes to the observed phenotypes. This kind of innovative screening technology allows scientists to target the cells that matter when developing immunotherapies
How to Detect Cancers Without Cell Biopsies?
However, there may remain complex challenges in immuno-oncology when it comes to cancers that make cell biopsies tricky or infeasible. Dr Pieter van der Velden, a Senior Researcher at Leiden University Medical Center, works on this problem in ophthalmology. He presented his and his team’s research in the same workshop as Dr Chien. Dr van der Velden’s research focuses on using cell-free DNA as a tool for quantifying lymphocytes in the eye. Cell-free DNA are nucleic acid fragments that naturally occur in the bloodstream; the Leiden University Medical Center team has used analysis of these fragments to yield generic markers for counting T and B lymphocytes.
- Understanding The Tumour Microenvironment With Advanced Technologies
- Perfecting Precision Medicine: The Promise of Biomarkers and Spatial Biology
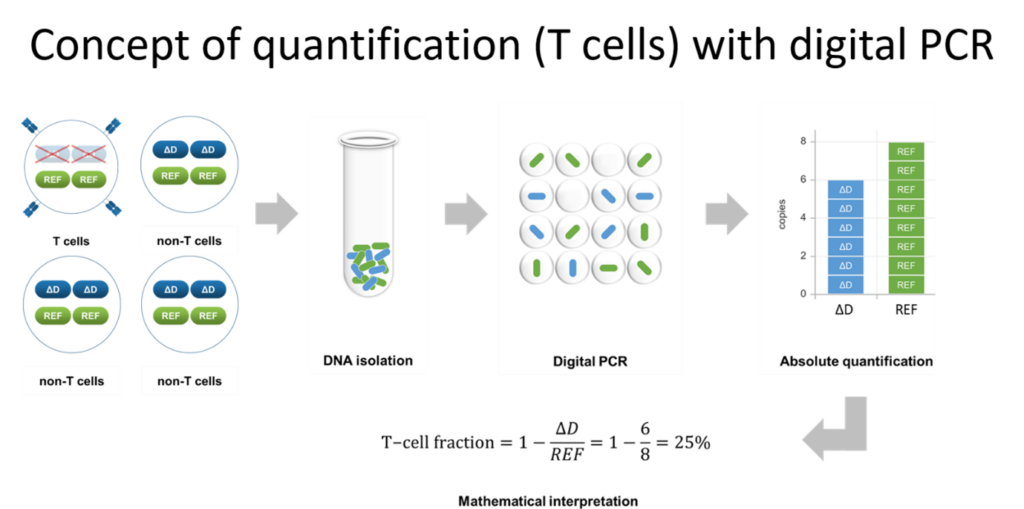
The Leiden scientists had discovered that at the IGH locus, the allele ?H becomes lost during B-cell maturation and therefore can be used as a marker to count the cells. But how do you count what is not there? “The answer is context”, Dr van der Velden explains, “Just like Dutch and Swiss cheese, the missing cheese in each cube determines the origin of the cheese.” By counting the loss of ?H alleles, the B-cell number can be quantified using a formula.
“Just like Dutch and Swiss cheese, the missing cheese in each cube determines the origin of the cheese.”

Dr van der Velden gave two examples of eye cancers in which cell screening is not a practical or effective option and where this technique would be advantageous. The first is lymphoma of the eye or vitreoretinal lymphoma. This particular cancer is tough to diagnose as its symptoms are very similar to ocular inflammation. Furthermore, the lymphoma cells are not stable in the anterior fluid, making them challenging for cell screening. Here, DNA based B-cell counting makes for an advantageous way to diagnose lymphoma in lieu of cell analysis.
The second example that Dr van der Velden gave was the case of uveal melanoma in the eye. Uveal melanoma presents as tumours that grow in the void of the eye and therefore requires the removal of the eye to access the tumour tissue for bulk analysis. Furthermore, gene expression profiles provide a rather weak quantification of T lymphocytes, making them a difficult to validate quantification method.
Here, DNA-based counting can be used to investigate markers for T-cells that indicate melanoma progression. ?B (?D) alleles at the TRB (TRD) locus are lost during T-cell maturation, just like ?H for B-cells, meaning that DNA-based counting methods are applicable to uveal melanoma with little need for intact cell samples.
The world of genomics and immunotherapy grows more exciting and encouraging by the day, so don’t miss out on the latest developments in immuno-oncology by signing up for our monthly Immuno newsletter! Also, consider seeing the experts in-person by attending Oxford Global’s next Immuno UK conference.