How DEL Screening is Used in Living Cells
DNA-Encoded Chemical Library (DEL) finding has been a landmark technology in the discovery of drugs, but scientists are eager to improve the efficiency and efficacy of the process. Higher and more reliable hit rates aim to improve DEL technologies, which would fast-track the drug discovery process even further than DEL screening has already taken it. Presenting at Oxford Global’s 2021 online Discovery week, Dr Iolanda Micco suggests that implementing DEL screening into living cells, rather than in plastic reaction tubes, represents one such valuable enhancement.
Why use Living Cells?
The advantage of using living cells in DEL screening is that they provide an environment that has physiological relevance to the target proteins studied, says Dr Micco. The cytosol of a living cell is composed of thousands of macro-molecules that just can’t be replicated in-vitro. So, using living cells to provide this relevant environment to the target protein would theoretically lead to lower attrition rates and higher success rates.
Another reason DEL scientists are attracted to working with living cells is to exploit them as a vehicle for target protein expression. As these proteins can be expressed directly in the cell, there is no need to purify or even handle the targets. Therefore, workflow is faster, easier, and more efficient allowing the screening of challenging targets.
The Challenges with Using Living Cells
But how do you get the DEL into a tiny living cell in the first place? Dr Micco’s answer is straightforward: inject it. African clawed frog (Xenopus laevis) oocytes are prime candidates for DEL injection due to their large size (about a millimetre in diameter), about 100,000 times bigger than somatic cells. The enormity of these cells makes them perfect for the microinjection of DELs. Furthermore, X. laevis oocytes are great candidates, as they are inexpensive, widely available, and known to be capable of expressing a wide variety of heterologous proteins.
A second challenge is that DEL members may bind to the endogenous cell proteins instead of the protein of interest, recording hits on irrelevant proteins. Dr Micco explained that the team’s solution is to label the proteins of interest with DNA. This DNA allows scientists to differentiate the protein of interest from the superfluous endogenous proteins after achieving hits.
Experiment, Data, and Outcomes
The experiment used cell Binder Trap Enrichment, which Vipergen is currently pioneering at its labs. In each experiment, the fusion mRNA of the protein of interest and ‘Prey’ was injected into the oocytes initiating them to express the fusion protein. When the protein is expressed, the ‘Prey’ protein is expressed fused to it, differentiating it from the endogenous proteins in the cell. The scientists then injected DELs into the oocytes along with a DNA encoded small molecule known to be receptive to the Prey protein, the ‘Bait’. The Prey DNA and the DNA barcode for the hit molecule then ligate into one bonded DNA strand (Fig. 1). Therefore, scientists will know whether a hit achieved is receptive to the protein of interest if its DNA barcode contains the Bait DNA when sequenced.
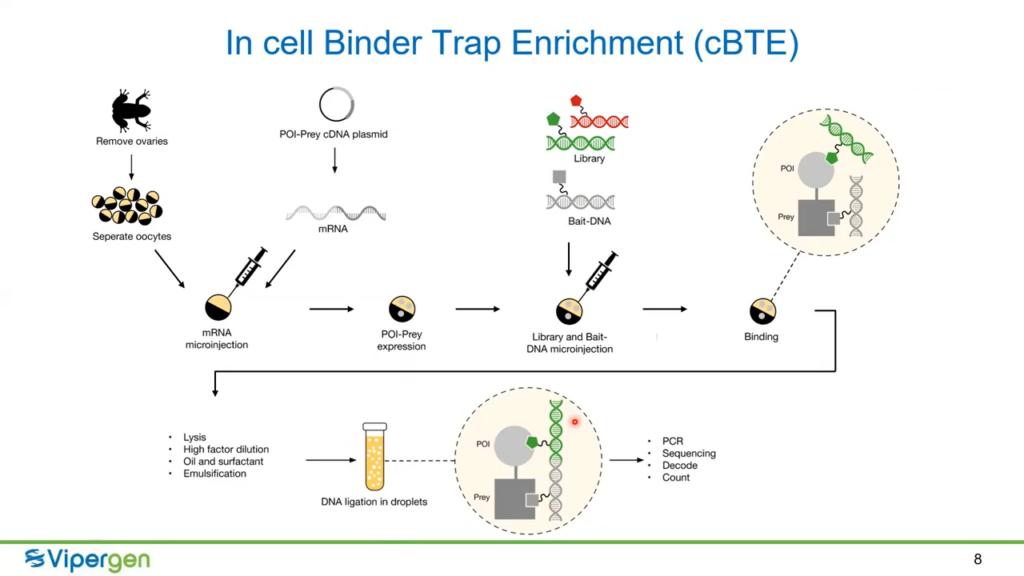
For the data to be collected, the scientists just need to amplify, sequence, and ultimately decode the DNA markers attached to the protein and Prey.
The team at Vipergen selected three proteins of interest to study DEL screening in living cells, these being p38?, ACSS2, and DOCK5. Each protein was experimented upon using three progressive concentrations of the Bait-DNA.
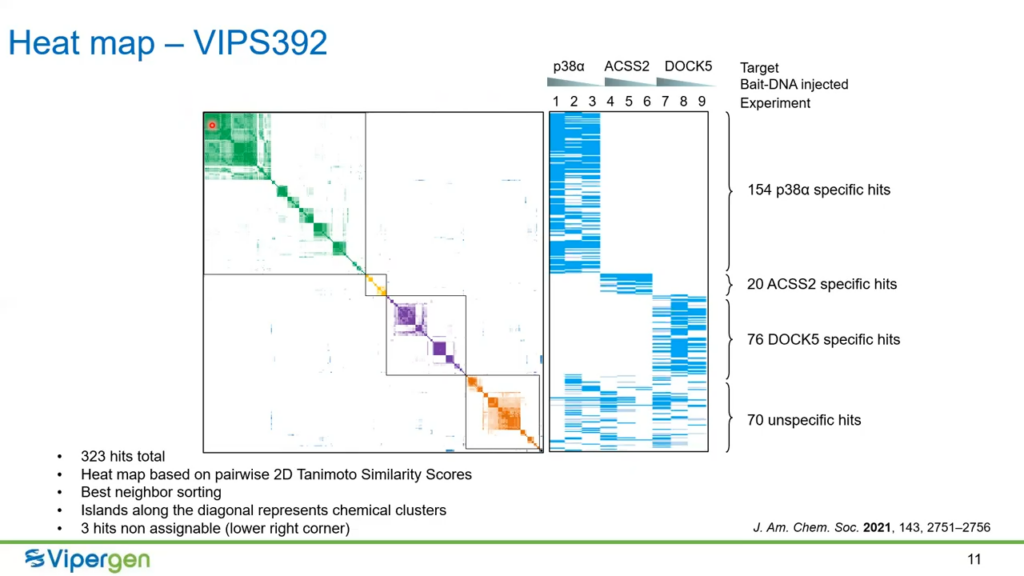
The organisation of this data in this way allows scientists to identify clusters of key compounds for further study and assay.
The most hit target protein, p38?, yielded 154 hits distributed across nine chemical clusters 21 were synthesised and assayed. They found 19 out of the 21 hits were shown to be inhibitors in an enzymatic assay.
Multiplexing, Orientation, and Competition
Dr Micco also talked about the technique’s potential for multiplexing, whereby two Prey mRNA are co-injected into the cell along with two Bait DNA. “Multiplexing is a very powerful approach. Not only does it save time and cost, but it also gives all the information on predicted specificity. So, for those applications where specificity is important, it is a very powerful tool.” Dr Micco explained.
They repeated the experiment in various ways to ensure the integrity of the data. The results were similar to the original findings in the following modes: both mono- and duplex-; using different Prey; using both N- and C- terminal fusion. Furthermore, their findings observed no crosstalk between Prey. “There is flexibility, freedom, and confidence with regards to the orientation of the proteins,” says Dr Micco.
Living Cells: a More Efficacious Environment
The techniques that Vipergen are using for DEL in living cells is up-and-coming. Using cBTE, they can screen for various potential targets: Human or pathogenic soluble proteins tolerating either N- or C- terminal fusions. However, there are some challenges for this technology. Integral membrane proteins can’t be screened in this way, and the team expects problems for screening proteins that bind strongly to DNA or could be toxic for the cell.
However, Vipergen can screen DELs under more relevant physiological conditions with this technology. DEL screening is, therefore, easier, and more efficacious than before, producing lower attrition rates. There is no need to purify the active target protein, which also means an expanded target space and a faster screening process. The experiments performed by Vipergen have shown that the technology is robust, creating reproducible results, and it shows that multiplexing is a viable use of the method.
To keep up to speed on the latest developments in DEL screening and the broader category of drug discovery, why not sign up for our monthly Discovery newsletter? Or consider coming along to our next Discovery Europe conference?