Achieving the Gold Standards of Organ Modelling: Complexity, Robustness, and Reproducibility

At Oxford Global’s Organ Modelling UK conference we were happy to host a great panel discussion bringing together key leaders and experts in the field of animal and disease modelling. The session was titled Overcoming Challenges in Complexity, Robustness and Reproducibility, and welcomed:
- Pelin Candarlioglu, Bioengineer/Cell Biologist, GlaxoSmithKline
- Lorena Benedetti, Scientific Investigator, GlaxoSmithKline
Issues in Disease Modelling
Moderator Mootaz M. Salman, Departmental Research Lecturer at the University of Oxford introduced the panellists and topics. The first topic to tackle was complexity: “How complex should your models be?” said Salman, “Academics like to incorporate lots of factors into their models, is that what big pharma needs? Or is simplicity the answer?”. Second, robustness: “how robust is your model?”, and finally, reproducibility: “What are the issues of reproducing these data?”
Salman said that one of the problems with complexity was that there was a lack of uniformity of definition: “What are organoids? How complex should our model be to call it an organoid? And what are organs-on-chips?” he asked Candarlioglu.
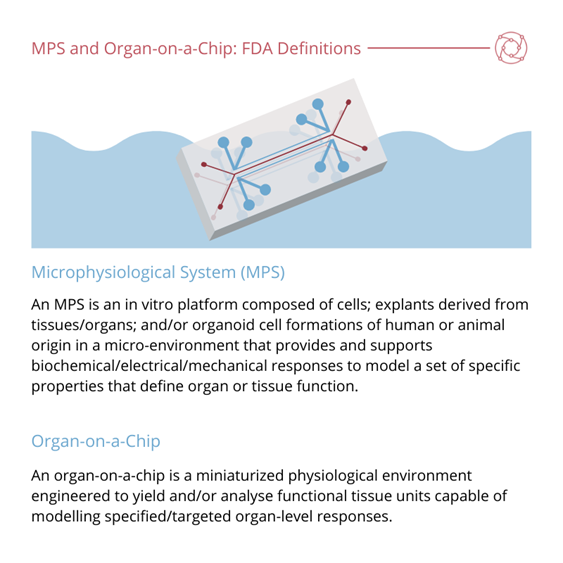
Candarlioglu commented on the great effort that has been put into the standardisation of the regulatory aspects of these models. She then discussed the current working definitions of organ-on-a-chip, organoid, and microphysiological system (MPS).
“The first attempt to bring about a definition for organ-on-a-chip was done by IQMPS [the Affiliate of the International Consortium for Innovation and Quality in Drug Development on MPS].” Their definition, said Candarlioglu, comprises the fact that organs-on-chips go beyond traditional 2D cultures by including the following design aspects:
- Multicellular environment within polymer or tissue drive metrics
- Mechanical cues such as stretch perfusion and breathing
- Incorporation of stem cells as drive cells
A problem with this definition according to Candarlioglu is that rather than defining the technology’s key elements, it is more of a list of properties. “IQMPS is working on an updated version of this which will be released quite soon,” Candarlioglu explained. She added that the FDA (with help from the IQMPS) have since created the definition that most people are familiar with at the moment.
One of the key elements of the FDA definition is that they have differentiated between microphysiological systems (MPS) and organ-on-a-chip – “which a lot of people until now were using interchangeably,” added Candarlioglu.
Along with this definition, Candarlioglu further mentioned the definition penned by the EU’s ORCHID project – an organisation which led to the formation of the European Organ on Chip Society (EUROoOCS).
An Organ-on-Chip (OoC) is a fit-for-purpose microfluidic device, containing living engineered organ substructures in a controlled microenvironment, that recapitulates one or more aspects of the organ’s dynamics, functionality and (patho)physiological response in vivo under real-time monitoring.
ORCHID definition of Organ-on-Chip
“So, what is the right level of complexity?” Candarlioglu tackled next. “An important question to ask is: is the target expression relevant to the in vivo?” For example, CAR Ts or TCRs require a certain amount of target expression for the right activation levels. “If you’re not able to get that target expression statically, you will need to use a mechanical stimulus to achieve that,” said Candarlioglu.
Defining Robustness
Salman then asked Benedetti the question of how to define robustness. Benedetti stated that it was important to understand which endpoint clearly recapitulated the aspect of the disease that researchers were setting out to model. “Once you have narrowed that down and found the right endpoint, you can make sure that you’re always ready to measure that endpoint robustly, whatever its level of complexity.”
- Using Organs-on-Chips to Catch Kidney Toxicity in Drug Discovery
- Challenges in Developing Organoids for Neuroscience
- Tackling Tumours In-Vitro with 3D Cell Cultures
Benedetti explained that she worked with mesenchymal stem cells differentiated into particular cell types. She wanted to make sure that they used the right medium to induce this differentiation. “Each vendor recommends a different passage window, but which is the right one for the phenotype you are looking for?”
Among the factors that Benedetti looks out for when making this decision are recapitulation of healthy tissue characteristics. Furthermore, the level of complexity needed to model the disease, and which conditions recapitulate the disease stage that they are trying to modify are also considered. “This is a very laborious process,” said Benedetti, adding that “it can take several years.”
Benedetti rounded off her section of the talk by stating that the use of publicly available transcriptomic datasets on the organ recapitulated inform on the identity and validity of the cells differentiated. This information alongside the use endpoint assays that reflect the physiology and functionality of the organ help building on the robustness of the model.
Want to hear more? Network with over 200 industry leaders at Discovery US: In-Person, where we will address the latest advancements in target identification, validation and HIT optimisation.
For more content on organ modelling, and other developments in the drug discovery sector, sign up to our Discovery newsletter, which will bring you content highlights, upcoming events, and more.