Single Cell Proteomics: Advancements and Potential

Our April 2022 Omics Series discussion group focused on the potential and future of single-cell proteomics. Oxford Global’s discussion groups bring together a select group of 15-20 key industry leaders for approximately one hour for in-depth knowledge sharing and conversation.
This month the group was led by Yann Abraham, Senior Principal Scientist at Janssen. In this position, Yann plays a key role in analysing and integrating datasets generated through partnerships with institutions in the French biomedical research ecosystem, leveraging synergies with internal projects from the Janssen Therapeutic Areas and initiatives from the Data Science team within DS Computational Sciences.
Abraham opens the group by answering, “why do we want to do proteomics?” He explains that “proteins are the building blocks of life, and most biology is driven by the amount, localisation and activation statuses of proteins. Conversely, diseases most likely occur when the proteins are incorrectly expressed, localised or activated. Therefore, understanding proteins is key to advancing biology and advancing the way we cure diseases.”
Single-cell proteomics allows you to do two equally important things to answer the questions posed by recent literature. First, they will enable you to document or study changes in composition. Second, Proteomics also allows you to examine changes in functionality, which is challenging to do with other methods.
Single Cell Proteomics Technology
One of the limiting factors of single-cell proteomics over the past few years has been the lack of efficient technology. Fortunately, this is changing fast. Abraham notes that you can “classify technologies by two axes: number of cells and number of proteins. So basically, when you’re doing immunohisto chemistry, you are doing single-cell proteomics.”
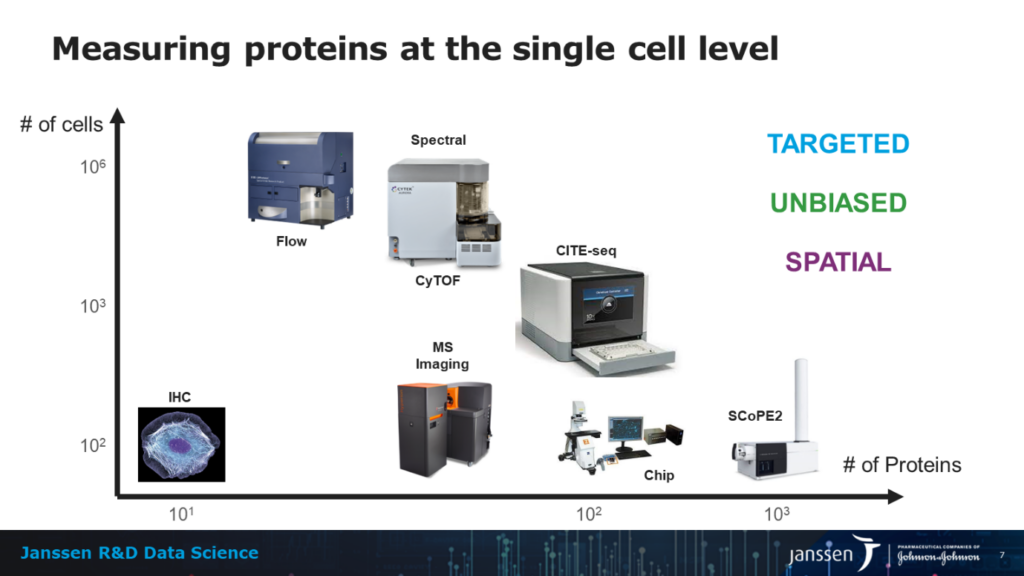
When discussing single-cell proteomics, many people will think about flow cytometry. This technology allows you to measure more markers on significantly more cells. CITE-seq is also a recent development which allows sequencing to detect proteins through DNA barcodes. Spectral cytometry combines the best of both worlds between flow cytometry and CITE-Seq. It can achieve similar flow heights as flow cytometry and an equal number of molecules as CITE-seq.
Now, there is a way to measure patterns in an unbiased way: mass spectrometry. Abraham explains that “recently, some people have pushed the boundary of what mass spectrometry can do by developing a technology to measure the protein content of a single cell at a reasonable scale. We’re still talking a few 100 cells at the time, we are not quite there yet, but the main advantage of this technology is that it’s unbiased.”
Abrahams elaborates that “all you need to do is a sequence of a protein and you can under certain circumstances detect the presence or the absence of particular proteins without the need of an antibody.” The most recent development for the field of single cell proteomics has been the so-called spatial technologies, either antibody based (Chip cytometry, Imaging Mass Cytometry) or unbiased (Image Mass Spectrometry), enabling deep profiling of tissues and tumors.
The Future of Single Cell Proteomics
Abraham highlights two significant developments he expects to see in the coming years. “There’s a technology called SELEX. Using the property of some RNA sequences to adopt 3D conformations that have protein-like properties, one can use in vitro evolution (repeated cycles of mutation and selection against a target) to raise a sequence that will recognize a particular target or even a particular conformation much in the way an antibody would. Such aptamers can be fine tuned to recognize specific isoforms, and detected either directly through sequencing, or by tagging them. So, one of the major advancements I see in the field is the extension of what you can target.”
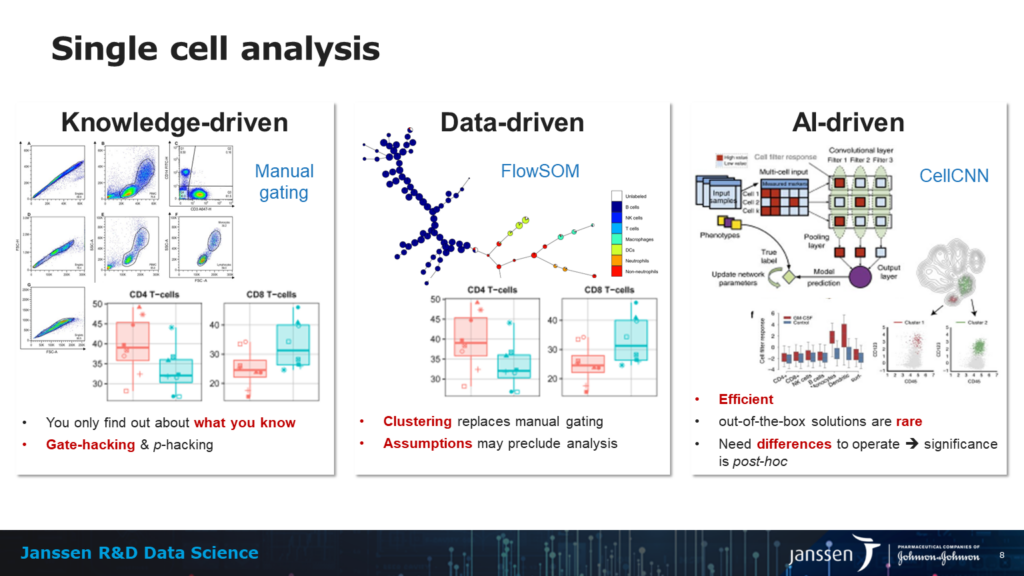
The other advancement is the field of AI-driven analysis. Abraham notes that “more and more people are moving towards data-driven solutions where one looks for differences at the single cell and not population level. Now there is a catch to this approach: it works under the assumptions that the difference you’re looking for is in the data you collected, and there is no guarantee of that. So, while I see AI-driven analysis as a major evolution in the future of this field, I think we will see it combined with descriptive views.”
Key Questions
One thing that makes Single Cell Proteomics difficult is the cost. Do you think this will change in the future?
I think that there are there two drivers to the costs. One of these is the price of the antibodies you’re putting in the mixture. For many of them, they are commodities, but then they are not necessarily cheap commodities. And for particular questions, companies can build to scale, so the price decreases. But sometimes, you want to add your own therapeutic antibody, and you want to know whether it’s binding the target and measure receptor occupancy. That can amp up the cost of your experiment. The second is reagents, be it sequencing reagents, in the case of cyclic or simply the heavy metals in the antibodies. So, unless we see a significant evolution in instrumentation, the limitations will be precisely that: the price of the antibodies and the price of the reagents.
What methods do you use for protein preservation?
So what we are dealing with nowadays is two solutions. Either we are using fresh frozen samples, or we are using fixated samples. In the latter case we are using custom panels where we have to do a lot of optimisation to make sure that all the antibodies we are interested in work. One of the big promises of CITE-seq was the possibility to monitor both surface protein as well as cytoplasmic proteins. Then it turned out that many antibodies didn’t work anymore after formalin-fixation.
Most available samples are paraffin-embedded. And that’s the killer question I usually ask any technology provider; is your technology compatible with paraffin-embedded tissue? A couple of years ago, the answer was no. You have some positive responses nowadays, but you may run into issues with some epitopes disappearing in the procedure. But it is much better than five years ago.
For more on single-cell proteomics sample preparation, read about Ryan Kelly’s work on sample miniaturisation and NanoPOTS.
Final Thoughts & Conclusion
At Oxford Global, we could not be more pleased with the turnout and feedback from this discussion. This meeting provided the perfect setting for exchanging ideas, sharing innovations, and discussing the ever-evolving digital therapeutics landscape. Sign up for our newsletter for the latest news on our Single Cell Proteomics Symposium, Omics US and Omics UK events for more on single-cell proteomics.






