Proteomics and Metabolomics: The Role of Cellular Microenvironment in Driving Tumour Progression and Metabolomic Fingerprint
Spatial proteomics is an invaluable tool for profiling the tumour microenvironment (TME), which plays a fundamental role in disease progression in organ tissue. Understanding how diseases operate at molecular, cellular, and tissue scales can abet treatment provision in proteomics and metabolomics. Additionally, real-world data (RWD) and real-world evidence (RWE) can be used to fill in gaps in research data to better predict the progression of disease in patients.
Single cancer cells frequently contain mutations in multiple genes, in addition to gross chromosomal abnormalities and widespread changes in their gene expression profiles. Epigenetic abnormalities leading to altered expression of these genes are also key for tumorigenesis: oncogenes and tumour suppressor genes are important not only for cell proliferation but also cell death determination. The hope is that unravelling the molecular circuitry of cancer cells will facilitate the better prediction of selective molecular targets for cancer therapy.
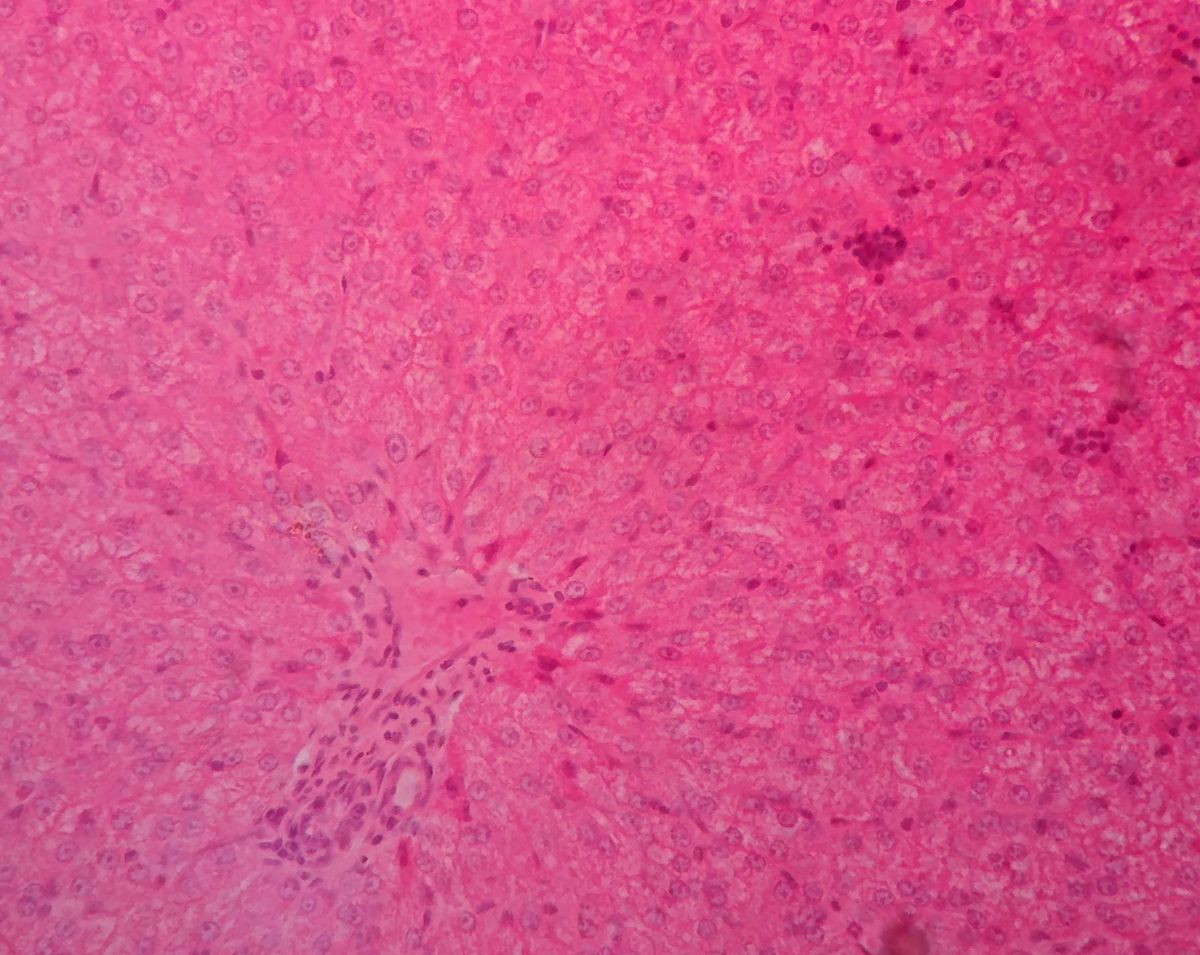
Spatial Metabolic Analysis for Investigating Liver Zonation
Mass spectrometry imaging is a vital tool for spatial proteomics, as it provides both spatial and highly dimensional spectral information. A common challenge in spatial metabolomics is the identification and annotation of metabolites. Metabolism is a fundamental aspect for every cell in the body, but precise metabolite identification and notation can be challenging for researchers.
One example of this is liver zonation, with the demonstration of spatial metabolic heterogeneity within a homogenous cell type. The liver has an interesting spatial heterogeneity which makes it particularly suitable for spatial metabolic studies. Liver zonation is verified with the analysis of spatial metabolic heterogeneity within a homogenous cell type.
Analysing liver tissue in this way through staining can reveal critical biological information about cellular state, property, and function. For example, chronic damage that stimulates fibrogenic processes may result in the deposition of excess matrix tissue and the impairment of liver functions. In the field of proteomics and metabolomics, there has been an identified lack of comprehensive analytical methods and integrative software tools for spatially resolved single-cell metabolomics.
To counter this, a research team from the University of Texas developed a spatial single nuclear metabolomics method (SEAM) scaling from in vitro cell cultures to heterogeneous clinical tissue sections. Without extra labelling, researchers can segment cells through using nuclear enriched metabolites: they can identify spatially heterogenous tissue and cell distribution patterns using metabolic profiles through integrative analysis.
Targeting Cancer Oncogenes in Proteomics and Metabolomics
Another key application of proteomic and metabolomic research is the interpretation of cancer oncogene behaviour. Until recently, there was a prevailing belief in the oncological field that if a cancer was dominated by a particular oncogene, then the way to kill that cancer was to target that oncogene. However, attempts to eliminate tumours by switching off the oncogene were only met with temporary success, with the same tumour recurring a few months further down the line.
The issue concerns the fact that tumours are highly heterogeneous: there is significant variation between tumour cells. As a result, focused action on specific death mechanisms renders therapies vulnerable to drug target alteration by somatic mutation. Over the course of treatment, efficacy declines and patients ultimately progress. Understanding and targeting de novo and acquired resistance remains a major obstacle to improving remission rates and achieving prolonged disease-free survival.
- How AI is transforming genomics
- Spatial omics and AI: an insight from King's College London
- On and off-target genome editing techniques
A central issue with cancer therapies is that tumour mutations drive a variation in behaviour — the treatment process acts as a selection pressure, creating tumours that are increasingly resistant to therapy. Focused action on specific death mechanisms renders therapies vulnerable to alteration of the drug target by somatic mutation.
Understanding and targeting de novo and acquired resistance remains a major obstacle to improving remission rates and achieving prolonged disease-free survival. Subsequently, many mutations drive a variation in behaviour. If a better understanding of tumours could be achieved, researchers may be able to improve their tracking of the course of treatment.
Heterogeneity at the Root of Recurrence in Proteomics and Metabolomics
Attempts to understand tumour behaviour by a research team at Stanford University revealed that resurgent tumour behaviour did not emerge solely from evolutionary dynamics. Attributes such as hypoxia, pH, glucose concentration, drug treatments, and secreted factors all influence the fitness of cancer cells and drive the evolution of a tumour.
In some ways, the condition of human tissue can be thought of as a river ecosystem. Different sections of a river will be suitable for certain organisms subject to the water’s temperature, turbidity, and salinity. The bloodstream of the body can be conceptualised in a similar way: forces like hypoxia can lead to changes in the local cellular environment, and may drive cancers to become extremely aggressive.
To evaluate tumour behaviour and response, researchers from Stanford replicated tumoral spatial architecture in a cell model, setting up nutrient gradients driven by blood vessels. The diffusion of nutrients out of those blood vessels was observed, and different cell types were observed to have different birth or death rates depending on their characteristics. From an initially mixed population of sensitive and resistant cells, the community evolved according to a multi-type stochastic birth and death process.
Multi-Dimensional Immunohistochemistry in the Study of Cell Microenvironment
In this study, the growth curves of different cells reflected a transition from a nutrient-rich regime to a nutrient-poor regime. Changes in growth rate purely driven by microenvironment may drive the formation of aggressive phages. Subsequently, novel imaging techniques such as multi-dimensional immunohistochemistry enable deeper investigations of molecular processes in context. Tumour microenvironments can drive tumour heterogeneity and evolution.
Across liver tissues and solid tumours, new spatial-based measures that capture the microenvironment are highly predictive of patient outcome in proteomics and metabolomics. There is a current need for experimental techniques to be coupled with sophisticated algorithms to analyse this data, and to extract novel features on the machine learning side.
Novel methods for spatial analysis are continuing to provide new insights into the state and function of tissue, enabling analysis of cell-to-cell interactions in the tumour microenvironment. To read more about new and ongoing research in spatial imaging, visit our Omics portal.







