An Overview of Liquid Biopsy: Non-Invasive Approaches to Monitoring Tumour Condition

Although the phrase ‘liquid biopsy’ has been around for many years, it was only introduced as a diagnostic concept in 2010 for the analysis of circulating tumour cells (CTCs) in the blood of cancer patients.
However, while most advances in understanding of liquid biopsy have come in the past 10-15 years, the first steps towards exploring the potential of cell-free DA for the genetic testing of cancer date back to 1948.
The present-day use of the term has now extended to the analysis of circulating tumour-derived vesicles.
In particular, it involves the analysis of circulating tumour DNA (ctDNA) and cell-free DNA (cfDNA), as well as extracellular vesicles (EVs), cell-free microRNAs (cfmiRNAs), and mRNA.
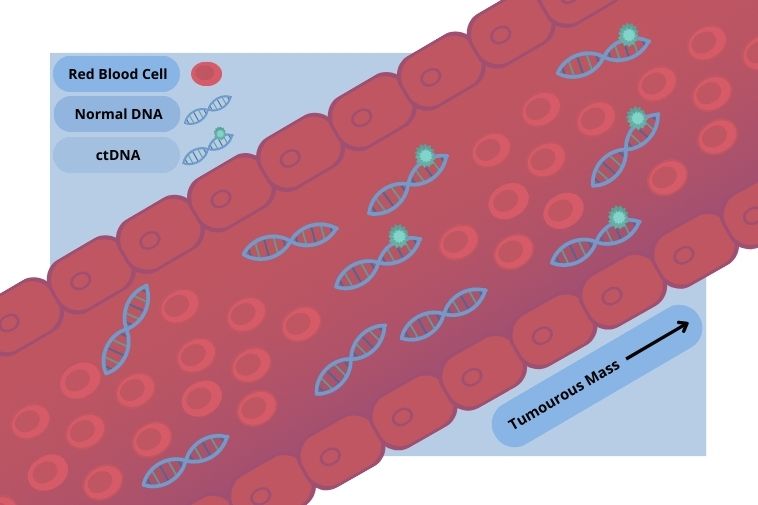
As single- or double-stranded DNA released by tumour cells into the blood, ctDNA harbours the mutations of its parent tumour, making it a good indicator of tumour progression in diseases such as non-small cell lung cancer.
One of the chief advantages of liquid biopsies is their value as a non-invasive alternative to surgical biopsy – they have the potential of revealing the molecular signatures of tumours to aid in the individualisation of treatments.
Although most studies to date have been performed on patients with carcinomas and melanomas, CTCs have also been detected in the peripheral blood of patients with primary brain tumours despite the presence of the blood-brain barrier (BBB).
Liquid biopsy assays are currently being validated for the early detection of cancer, with the aim of reducing cancer-related mortality.
However, the use of liquid biopsy-based detection of the early stages of solid cancers remains a challenge.
New blood-based biomarkers are currently being validated in clinical trials, including miRNAs, exosomes, and tumour-educated platelets.
Technologies for the Detection of ctDNA and cfDNA
Several different approaches exist for the detection of ctDNA and cfDNA in liquid biopsy analysis. Each has their own advantages and disadvantages which may lend themselves to a particular use case.
Droplet Digital Polymerase Chain Reaction (ddPCR)
The detection rate of ddPCR for genomic material can be as low as 0.01 to 0.1%, and the approach is useful for identifying potentially rare mutations, as well as calculating copy number variants. However, it can only be used to evaluate the presence of characterised sequences.
Beads, Emulsion, Amplification, and Magnetics (BEAMing)
Similarly to ddPCR, BEAMing is a relatively sensitive and inexpensive method of screening for known mutations. It combines PCR with flow cytometry, and can detect alterations in genomic material at concentrations as low as 0.01%, with excellent concordance to tissue testing.
Cancer Personalised Profiling by Deep Sequencing (CAPP-Seq)
CAPP-Seq identifies alterations in ctDNA and cfDNA by utilising large genomic libraries and individual patient sample sequence signatures. It can identify major mutation types such as insertions and copy variants, but it cannot identify fusions.
Tagged Amplicon Deep Sequencing (TAm-Seq)
TAm-Seq allows for a very specific and sensitive analysis of approximately 97% and can detect DNA levels as low as 2% by using primers to tag and identify genomic sequences. It has a high sequencing flux, a reduced sequencing time and cost, and can simultaneously sequence millions of DNA molecules.
Whole Exome Sequencing (WES)
Whole exome sequencing provides characterisation and analysis of all present tumour mutations, meaning it can be used to identify potential oncogenes and tumour suppressor genes. However, its sensitivity may be lower than other methods as it includes exomic alterations.
Whole Genome Sequencing (WGS)
Whole genome sequencing evaluates the entire tumour genome to determine characterised and deleterious alterations, along with variants of unknown significance. It has great potential for a comprehensive evaluation of all tumour mutations, but is limited by quality assurance, ethical issues, time, and cost.
Whole Genome Bisulfite Sequencing (WGBS-Seq)
WGBS-Seq is the gold standard in DNA methylation analysis: it provides a single cytosine measurement and has a high degree of accuracy. While it can discover partially methylated domains in cancer cells, DNA must exist in varying degrees of degradation. As such, this method may have reduced sensitivity.
How do Liquid Biopsies Supplement Diagnostic Approaches?
Solid biopsies are still the gold standard for tumour diagnosis, but they only represent a static and spatially constrained representation of tumour condition from the time of surgery.
Liquid biopsy approaches offer a more holistic assessment of tumour condition and state, allowing for continuous monitoring of cancer state and spread.
- dPCR Market Trends and Predictions for Future Development
- Exploring the Progress of PCR Technology and Novel Applications
- Solving Challenges in Cancer Detection Using Droplet-Based PCR
Liquid biopsies also avoid significant clinical risks such as wounding or infection that accompany solid tumour biopsies, which can be a particular concern for immunosuppressed patients.
While liquid biopsy is unlikely to fully replace or directly compete with solid biopsy as a diagnostic approach in the near future, some predict that liquid biopsy will be fully integrated alongside solid biopsies in the clinic in the next five years.
Get your weekly dose of industry news and announcements here, or head over to our Omics portal to catch up with the latest advances in tumour analysis. To learn more about our upcoming NextGen Omics UK conference in London, click here to download an agenda or register your interest.
Related Resources






